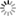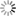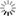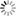Interview with Nobuyuki Matubayasi and Shun Sakuraba, developers of ERmod
|
Inteviewees: Nobuyuki MatubayasiAssociate Professor,
|
 |
|
Interviewer: Shingo YonezawaAssistant Professor, |
Materials are all around us, and we use various functions of these materials. For example, polymers that are used in diapers have the function of adsorbing and releasing water molecules. The positive and negative electrodes of a battery have the function of drawing in and releasing electrons. Then, is it possible to calculate such various functions of materials within a single framework and even to predict unknown functions? In this section, we will introduce the software application ERmod (prouounced "eee-arr-mahd") that has been developed to address this question.
"Dissolving" and "Mixing": Material Functions From a Chemical Approach
The original developer of ERmod, Nobuyuki Matubayasi, had an idea: if we expand the concepts of "dissolving" and "mixing," it may be possible to achieve a unified understanding of material functions. In the example of the water-absorbing polymer, if we view the water molecules as a "solute" and the polymer̶the environment that takes in these water molecules̶as a "solvent," then the phenomenon of water molecules being taken in by the polymer can be thought of as a "solute" (water molecules) being dissolved in a "solvent" (polymer). Similarly, in chemical reactions (reduction reactions) in which electrons are taken in, we can think of the electrons as being "dissolved" into the molecules. This approach is not just paraphrasing; it gives rise to tremendous advantages.
The "solution theory," which links these macroscopic "dissolving" and "mixing" phenomena to microscopic molecular-level dynamics, makes it possible to conduct high-speed calculations that are difficult simply by tracking movements of individual particles. From this bold shift in perception was born the software program ERmod.
The Birth of ERmod
Matubayasi was originally an experimental chemist who was studying liquids. "I wanted to combine experiments with calculations in order to pursue research both safely and efficiently," he thought. So he began to conduct simulations using the molecular dynamics (MD) method. The MD method is a calculation technique that tracks movements of individual atoms and molecules over time. However, with this method, it was often difficult to obtain results that were truly useful for experiments.
Matubayasi wanted to develop a calculation method that could not only reproduce experimental results but also predict unknown facts. This desire was the starting point for the development of ERmod.
The gold-standard way to compare experiments and calculations is to calculate the quantity called "free energy." The free energy is an indicator of the overall stability of aggregations of molecules or atoms. The free energy is fundamentally important because it determines how the aggregations experience various changes, such as dissolution and chemical reactions. However, it takes an extremely long time to calculate the free energy using only the MD method.
As shown in the upper half of Fig. 1, this is because to do this it is necessary to calculate not just the state before the change occurs (initial state) and the state after the change is completed (final state) but also numerous intermediate states.
Matubayasi hit on the idea of using the solution theory by which we can obtain the free energy from the molecular-level information of the system. According to the solution theory, it is possible to construct a function that takes a histogram of molecules on a certain parameter (such histograms are known as a distribution function) as an argument. Because this function is a function of a function, it is called a "functional." From this functional, the free energy can be calculated. In other words, by using the MD method only to calculate histograms at the initial and final states and entering these histograms into an appropriate functional, we obtain the change in free energy. Thus, one can avoid all the time-consuming calculations for the intermediate states and calculate the free energy change in a short period of time (the bottom half of Fig.1).
 |
Fig.1 Pattern diagrams showing a comparison between the functional theory in energy representation (the basis for the ERmod algorithm; lower half) and a conventional calculation method (upper half). ERmod uses the solution theory with a distribution function to achieve high-speed calculation. |
The next question is what kind of histogram (distribution function) should be used. The first that comes to mind is the distribution function on positional information. However, it is not evident which real-space parameter should be used among the enormous quantity of positional information (even for the simplest case, for a system of N molecules, there are 3N parameters). One day Matubayasi, who had been wrestling with this question, came up with the idea of using the distribution function for the interaction energy between molecules. It was an exciting moment. "I had just awakened from sleep when the idea popped into my head, and then it was just a matter of writing down the equations." Furthermore, he realized that the use of the energy distribution function provides several advantages, such as the ability to handle nonuniform systems and systems with degrees of freedom within molecules. This "Columbus' egg" type of inspiration (something that looks easy only after it has been achieved) was greeted with astonishment. The comment of a certain famous professor is still fresh in Matubayasi's mind. "Was it really you who came up with this idea?" he asked in surprise.
ERmod represents this idea turned into an actual program. By the end of the 2000s, ERmod had been incorporated into various MD calculation programs as a module. The name "ERmod" is a combination of the initials for "Energy Representation" and "mod," meaning module. So it retains a vestige of its early days as a module.
Development as a Standalone Program
The major turning point for ERmod came when Shun Sakuraba became involved in its development. Sakuraba was a software developer who had previously worked at an IT venture firm, and he was also a theoretical chemist who had previously performed calculations for biomacromolecules. "I realized that, with the rapid diversification of computers and MD calculation methods in recent years, it was going to be difficult for ERmod to continue to be developed as a module in MD software programs," he says. So Sakuraba took on the major task of converting ERmod from a module into a standalone program. First of all, the program, originally written only in Fortran, was modified using C and Python to strengthen its cooperability with other MD programs.
Parallelization and other improvements were also added to the core computing portion of the program. As a result, the calculation speed increased by thousand times. He also upgraded the documentation, improved error messages, and made it possible to run the program easily in a variety of environments.
In this way, a great deal of effort was paid to such behind-the-scenes works that are designed to make the program more convenient for users. Moreover, in order to expand the user base, they published the entire program including the source code on the Internet. The program is now being downloaded by users around the world.
"Almost all of my proposals were accepted by Prof. Matubayasi̶in fact, to a worrying degree," says Sakuraba. Matubayasi also says that his policy is to begin a joint research project with trusting the partner by 100%. I felt that their relationship of trust was the driving force for the great leap of ERmod.
The Effect of "Selection and Focus" in ERmod
ERmod's distinctive feature can be described in a single phrase: "selection and focus." In a sense, it ignores all the vast information in the real space and focuses on highspeed calculation of the free energy, which is critical for experiments. This is a reflection of the development philosophy that Matubayasi has had since the very beginning: the desire to create a program that is useful for experiments. The program is also characteristic for its use of other MD programs to calculate the distribution functions.
Naturally, there are also other ways to calculate distribution functions independently, repeating the process over and over again until the error converges. But ERmod outsources this distribution-function calculation to an existing MD calculation program, and concentrates on achieving more advanced functions. Importantly, there is a theoretical background ensuring that this way does not matter: Due to the variational principle, errors in MD calculations are not heavily reflected in the result of the free energy calculations. The effect of "selection and focus" in ERmod is evident in the actual calculation results.
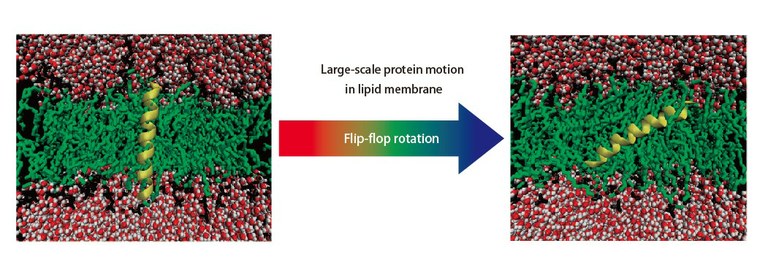
For example, when calculating the change in the free energy before and after a solute is dissolved in a solvent, the required time for calculation is less by several dozen times as compared to the amount of time required for the ordinary method (rigorous calculation), with a degree of accuracy that is by no means inferior. ERmod is currently being used in joint research with Toray Industries, Inc., for example, to design water-absorbing polymers (Read p.7 for more details on this joint project). The program has also achieved a wide variety of applications as exemplified in Fig.2.
|
|
| Fig. 2 Example of ERmod application. Pathway of large-scale protein motion in lipid membrane. In addition to this example, ERmod has been applied to a variety of systems. |
Evaluation for Behind-the-Scenes Work
"The lack of detailed documentation is one of the major reasons that Japanese academic software applications do not often become world-famous," says Sakuraba. He also displays a professional's perspective in saying that error messages are not only important for users. "From a long-term perspective, improving error messages also eases the burden on the developers." However, no methods have been established to provide proper evaluation of such tasks.
Matubayasi and Sakuraba are united in saying that this is a problem. I myself also have experiences to develop programs for my experiments. Thus, I understand that it is very annoying to compile documentation explaining the program and provide a complete array of error messages. And yet despite this annoyance, such efforts are not often evaluated properly. Similar types of work for experimental laboratories are arranging shelves or processing wastes. How should this type of behind-the-scenes works be evaluated properly as an important part of a researcher's job? How should we avoid a case of "the earnest man makes a fool of himself?" I get a feeling that this is becoming a serious issue not only in academic software application development but in various other fields of research as well.
The Future of ERmod
How will ERmod be developed in the future? The first objective is to extend its use to large-scale computers. For example, the way to use ERmod on the K computer has been comprehensively introduced in the ERmod website. The developers also intend that the program be installed on large-scale computers so that users can use it without compiling and installing by themselves. If this is realized, users who are not familiar with theoretical chemistry can more easily access ERmod, leading to applications in a wider range of systems such as biological materials and batteries.
Moreover, progresses in the underlying theories will also be indispensable for further development of ERmod. For example, if time evolution of a system can be calculated, the range of applications will become even wider. It will be possible to determine the lifetime of the association state of a certain molecule, and to calculate electrical conductivity. Matubayasi is also planning to incorporate the "coarse graining" technique in order to perform calculations for even larger targets. The “coarse graining” is a technique to increase the speed of calculation by, for example, considering a polymer of 100 monomers as a group of 10 clusters, each consisting of 10 monomers. Such progress would undoubtedly further expand the range of applications for ERmod.
ERmod has been progressed through the harmony of Matubayasi's insight and Sakuraba's software development expertise. I am looking forward to see further developments and success in the future.
Nobuyuki MatsubayashiAfter receiving his Ph.D from Rutgers University, he joined the Institute for Chemical Research, Kyoto University, where he is currently Associate Professor. Originally he conducted experimental research into supercritical water, but shifted his focus to theoretical research. He has received many awards, including the Young Researcher's Award from the Minister of Education, Culture, Sports, Science and Technology, for the formulation of a functional theory in energy representation (the basis for ERmod). He is also a proud father who decorates his laboratory with many creations made by his beloved daughters in kindergarten and elementary school. |
 |
 |
Shun SakurabaHe received his doctorate in computational biology from the University of Tokyo Graduate School of Frontier Science. He conducted research into protein simulation. He is a skilled programmer who was involved in a venture firm (established by a classmate while they were studying at the University of Tokyo) and worked on the development of a compiler. He is currently pursuing research as a postdoctoral fellow at the Japan Atomic Energy Agency, and is also playing a leading role in ERmod development. |
|
◆Interviewer's Postscript◆ Shingo YonezawaI am an experimental physicist mainly studying uperconductivity, and thus I am not an expert of theoretical chemistry. In this interview, however, both Prof. Matubayasi and Dr. Sakuraba explained their study in great depth and spoke passionately about the subject. So I came to understand their subject very clearly, and the scheduled interview time was up before I knew it. I feel that Prof. Matubayasi, who seems very easygoing, and Dr. Sakuraba, who seems very earnest, make an excellent team with a very good harmony of their personalities. |
 |