Modylas Developer : Yoshimichi Andoh
Modylas, a program that can calculate the trajectories of 10 million atoms simultaneously, will soon be a reality. We talked to developer Yoshimichi Andoh, a researcher at the Susumu Okazaki Laboratory at Nagoya University who is playing a central role in the development project, to ask what kinds of things Modylas can tell us and what the applications for the software will be.

What kind of program is Modylas?
I liked the name Modylas, so I began by asking where it came from. "Modylas" stands for MOlecular DYnamics simulation software for LArge System.
In molecular dynamics (MD) calculations, the initial coordinates of all of the atoms making up the groups of molecules (systems), as
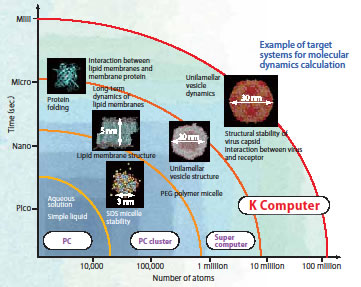
well as a variety of force-field parameters describing the interactions between atoms, are entered, and then, the Newtonian equations of motion for the all atoms are numerically solved in order to analyze the structure and changes (dynamics) of the system. Since calculating the interactions between atoms requires an enormous number of calculations, the scale of the systems that can be handled has increased with progress in computers. However, existing supercomputers can only handle systems that are several hundred thousand to several million atoms in size (Fig. 1). Yoshimichi Andoh is a specialist in molecular dynamics. He is both well-versed in computer science and a skilled computer programmer. But even for him, it was not easy to develop software for an advanced supercomputer.
"The K Computer is a complex computer made up of more than 80,000 computing nodes. The key to bringing out the maximum capabilities of the K Computer is the construction of algorithms that reduce the number of communications, enabling the computer to calculate efficiently. Developing Modylas often required the most advanced technologies that only a specialist in computer science would understand. Up to now, for crucial sections I had pursued research jointly with a software development engineer (from Fujitsu). But in order to conduct joint research smoothly, it was necessary to have a good understanding of computers. And computer science is a field in which new technical terms are being produced all the time. So we sometimes had trouble communicating." The biggest problem was how to efficiently incorporate the Coulomb force (electrostatic force), one of the forces that acts between atoms, into the MD calculations. The Coulomb force operates to a far enough distance that it can even be felt by human beings. As the Coulomb force is thought to control phenomena in the micro world at the atomic level as well, naturally it must be accurately included in simulations.
"On supercomputers up to now, the Particle-Mesh Ewald (PME) method has been used to calculate the Coulomb force.
This is an approximation calculation method that employs the fast Fourier transformation (FFT). Although it was sufficiently precise, there was a problem in that, as the number of calculation nodes increases, the traffic of communications between nodes resulting from the FFT becomes enormous, and this decreases calculation efficiency. In the case of the K Computer, which will consist of more than 80,000 nodes, a more efficient method of calculating the Coulomb force will have to be used in place of the PME method. Accordingly, we developed a new program for calculating the Coulomb force, based on the Fast Multipole Method (FMM). With the FMM, the force from nearby particles is calculated precisely, while the contribution from more distant particles is treated as a group. This results in an algorithm that is capable of high-speed simulation while still preserving a high level of precision (Fig. 2). Nevertheless, as the program is far more complex in structure than the PME method, it has almost never been used in actual MD software. We're the first to implement it in MD software on the assumption that it will run on a supercomputer made up of several tens of thousands of nodes . " The implementation of FMM is now the biggest selling point of Modylas.
|
|
Fig. 1 Size and time scale of the systems that are the subject of molecular dynamics (MD) calculations. The scale of calculations has expanded along with progress in computers and algorithms. The first attempt at MD calculations, in 1957, was performed for a system consisting of only 100 atoms. |
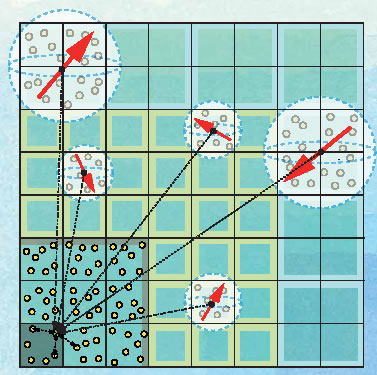 |
Fig. 2 Calculation of the Coulomb interaction using the FMM |
What can Modylas tell us?
So what kind of simulations will be possible using Modylas? Modylas can conduct molecular dynamics calculations for systems made up of 10 million atoms, which would take 500 years on existing supercomputers. The origins of the structural stability of biological macromolecular aggregates in the body such as viruses is a topic that is of great interest to scientists. Moreover, highly efficient MD calculation software will be useful for a wide variety of nanoscale research areas, such as predicting protein structures and elucidating the origin of the structure change in cell membranes caused by some diseases.
Dr. Andoh says that another of the pluses of this research is being able to see clear images in simulations. On the day of the interview, he showed me an actual image of a virus particle in water, and I was able to see immediately that it had a very interesting structure. The shell of the virus, made of a protein called a capsid, takes a certain form in which the same parts of the protein clump together.
"By performing calculations on the K computer, it will be possible to track the behavior of all of the approximately 10 million atoms, ranging from the atoms that make up the surrounding water molecules to the atoms that make up the virus. Up to now, we have not clearly understood in scientific terms why viruses become stable when they form this structure. I want to solve this riddle at the atomic level through simulations on the supercomputer."
What are the applications for Modylas?
A great deal of attention has focused on applications for Modylas in the area of drug discovery. It is difficult to determine through experimentation how viruses infect cells, or the molecular mechanism of the response between viruses and immune systems. Researchers hope that all-atom MD simulations will become a new method that can be used instead.
"By quantifying the interaction between a virus and a receptor as free energy, we will be able to clarify, at the molecular level, the reason that a virus attaches itself to a specific receptor in the human body. And if we make it so a different molecule with a similar structure attaches itself to the connector on the virus before the virus attaches itself to the receptor, we'll be able to block the virus from intruding into the interior of the cell. Understanding the structure of a virus will also lead to the development of drugs that can block that virus."
"In addition, this is also expected to be able to contribute to the development of drug delivery systems (DDS) that use the vesicles, which are matter transport carriers within cells. Vesicles are bag-like structures with the aqueous phase enclosed in a lipid bilayer membrane. The drug is injected into the interior of the vesicle, or into the lipid bilayer membrane, to deliver it efficiently to the affected area in the body. In this DDS development research, we still don't have a clear understanding of basic matters such as the interaction between the lipid membranes making up the vesicle and the encapsulated drug, or the interaction between the vesicle and the cell membrane that is the target of membrane fusion. So new knowledge obtained through MD simulation is needed."
Currently the calculation efficiency is poor for non-dense systems such as foam formation and the formation of stars in outer space, so these systems appear to be not really suitable for calculation by Modylas. Modylas is particularly good at calculating systems that are filled with materials and have a generally uniform density, as in the case of simulations of viruses in which the water and the target protein are entered and calculations are performed. As long as there is a good idea, Dr. Andoh hopes Modylas will be used for a variety of targets regardless of whether or not they are biomolecules.
"I want to lead the world with unique software developed in Japan"
There are not many researchers like Dr. Andoh, who combines a basic knowledge of physics and chemistry with programming skills.
"Currently the researchers in the molecular dynamics field who do both MD code programming and research are a minority. Ideally I'd like them to become a majority." There is also a great significance in passing on essential knowledge in Japanese software development to the next generation. "Currently the molecular dynamics community in Japan does not have software that can be used by anyone and is capable of highly efficient calculations that put it on par with the rest of the world. In contrast, the U. S. and Europe began pursuing software development around 20 years ago through joint research projects that brought together specialists in various fields such as computer science and physics. So in terms of organizational capabilities, Japan is quite a bit behind. Naturally it's possible to import software from the United States and use that. But we would only be using the software, and my fear is that through successive generations we would gradually lose an understanding of the basic principles that form the essence of the software. This could be said about any field. But in order to prevent this kind of hollowing-out of knowledge, it's crucial that we start from scratch to create unique state-of-the-art software in Japan."
From this point on, researchers in molecular dynamics and various other fields with specialist knowledge will come together to begin the final tuning of Modylas on the K Computer. Bringing together specialists from different fields will undoubtedly result in a definite increase in the calculation speed of each of the software parts. In this process, Dr. Andoh will continue to serve as the "captain" of the Modylas project and work to optimize overall performance.
"Up to now, we've resolved the difference in terminology among different fields through close communication. In the future as well, we plan to work together to bring the project to a successful conclusion." The goal is to create a program that reaches the top level on the world stage.
 |
Andoh Yoshimichi |
 |
Yoshimi Kubota |
【Dr. Andoh as seen from an interviewer's perspective】
At 5:00 p. m. on March 11, the day of the Great East Japan Earthquake, Dr. Andoh sent us the following email: "In this type of emergency, it would be especially difficult for Ms. Kubota to travel to Nagoya. So I think it would be a good idea if we postponed tomorrow's interview." As a result, the interview that had been scheduled for the 12th was postponed.
This solicitude reflects Dr. Andoh's character. The care he takes in his research activities needs no introduction, but the solicitude he shows toward joint researchers and the way he takes care of his juniors are also hallmarks of his character. In simulations for the K Computer, he not only takes the lead in pursuing software development but also plays a crucial role in establishing communication among specialists. Dr. Andoh himself is conscious of his role as the "captain" of the Modylas project team.